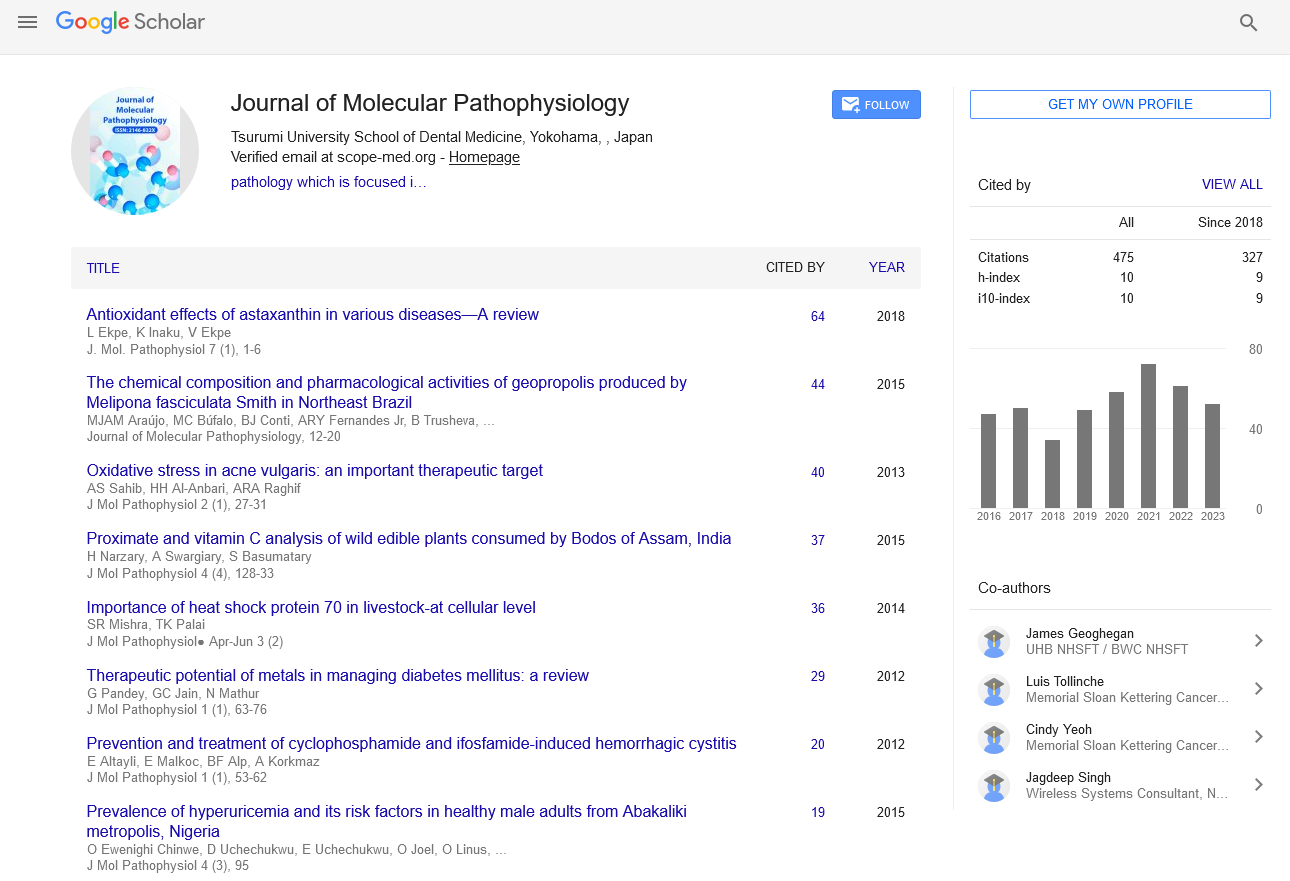
Editorial - Journal of Molecular Pathophysiology (2021)
SARS-CoV-2 Structure and Pathophysiology
Srinivas M*Srinivas M, Department of Orthopedics, Harvard Medical School Clinical Research Training, Egypt, Email: Srinivasmutthi@gmail.com
Published: 28-May-2021
To understand the pathogenic mechanisms of SARS-CoV-2 and to discuss the current therapeutic targets; it is important to describe the viral structure, genome, and replication cycle. CoVs are positive-stranded RNA viruses with a nucleocapsid and envelope.
A SARS-CoV-2 virion is approximately 50–200 nm in diameter and has a +ssRNA genome of approximately 29.9 kb in length—the largest known RNA virus with a 5′-cap structure and 3′- poly-A-tail and possess 14 putative open reading frames (ORFs) encoding 27 proteins The virion has four structural proteins, known as the S (spike), E (envelope), M (membrane), and N (nucleocapsid) proteins; the N protein holds the RNA genome, and the S, E, and M proteins together create the viral envelope.
The spike glycoprotein-S facilitates the virus attachment to the angiotensin- converting enzyme 2 (ACE2) receptor and fuses with the membrane of the host cell. SARS-CoV-2 then uses serine proteases TMPRSS2 (transmembrane protease serine 2) for S protein priming, infecting the target cells. The spike proteins of SARS-CoV-2 contains two subunits; S1 receptor binding subunit and S2 fusion subunit, to mediate the virion binding to receptor protein and initiate membrane fusion. The S1 and S2 subunits are divided by the S cleavage site. To facilitate virion attachment to receptor and fuses with cells membrane, the spike protein needs to be cleaved by cellular proteases from the S1/S2 cleavage site. Interestingly, the molecular analysis of S proteins identified an insertion at S1/S2 site, which is absent in other SARS-CoV, though the importance of this insertion is still unknown, it seems that this unique insertion is providing a gain-of-function advantage for an easy cell infection and efficient spreading throughout the human host.
The viral RNA hijacks the host cell’s machinery to initiate the viral genome replication and polypeptides chain synthesis and form the replication-transcription complex (RCT) needed to synthesize the sub-genomic RNAs as well as structural proteins (envelope and nucleocapsid).
The viral envelope has a crucial role in the viral assembly, release, and promoting viral pathogenesis However, the exact role of the many small viral peptides (e.g., those of glycoprotein subunits) has not yet been described. More research is needed to understand the structural characteristics of SARSCoV- 2 that underlie various pathogenic mechanisms.
Symptoms of SARS-CoV-2 resemble those of the common cold, including fever, coughing, and shortness of breath . However, the infection can lead to pneumonia, multi-organs failure, severe acute respiratory syndrome, and even death in severe cases. Elderly individuals (aged > 60 years) and people with underlying chronic health conditions are more susceptible to severe disease (18.5%) as compared to children and younger healthy adults (6%). The clinical data collected from the nonsurvivors patients revealed that the most distinctive comorbidities of SARS-CoV-2 infection were hypertension (24–75%) and diabetes mellitus (16.2– 35%). Notably, the most frequent comorbidities were reported in SARS-CoV-2 patients treated with angiotensin-converting enzyme (ACE) inhibitors. SARS-CoV-2 binds to host cells through the ACE2 receptor, which is expressed by epithelial cells of the lungs, intestines, kidneys, brain, and blood vessels . The expression of ACE2 is substantially increased in diabetic and hypertensive patients, treated with ACE inhibitors and angiotensin II type-I receptor blockers (ARBs), which consequently promotes SARS-CoV-2 infection severity.