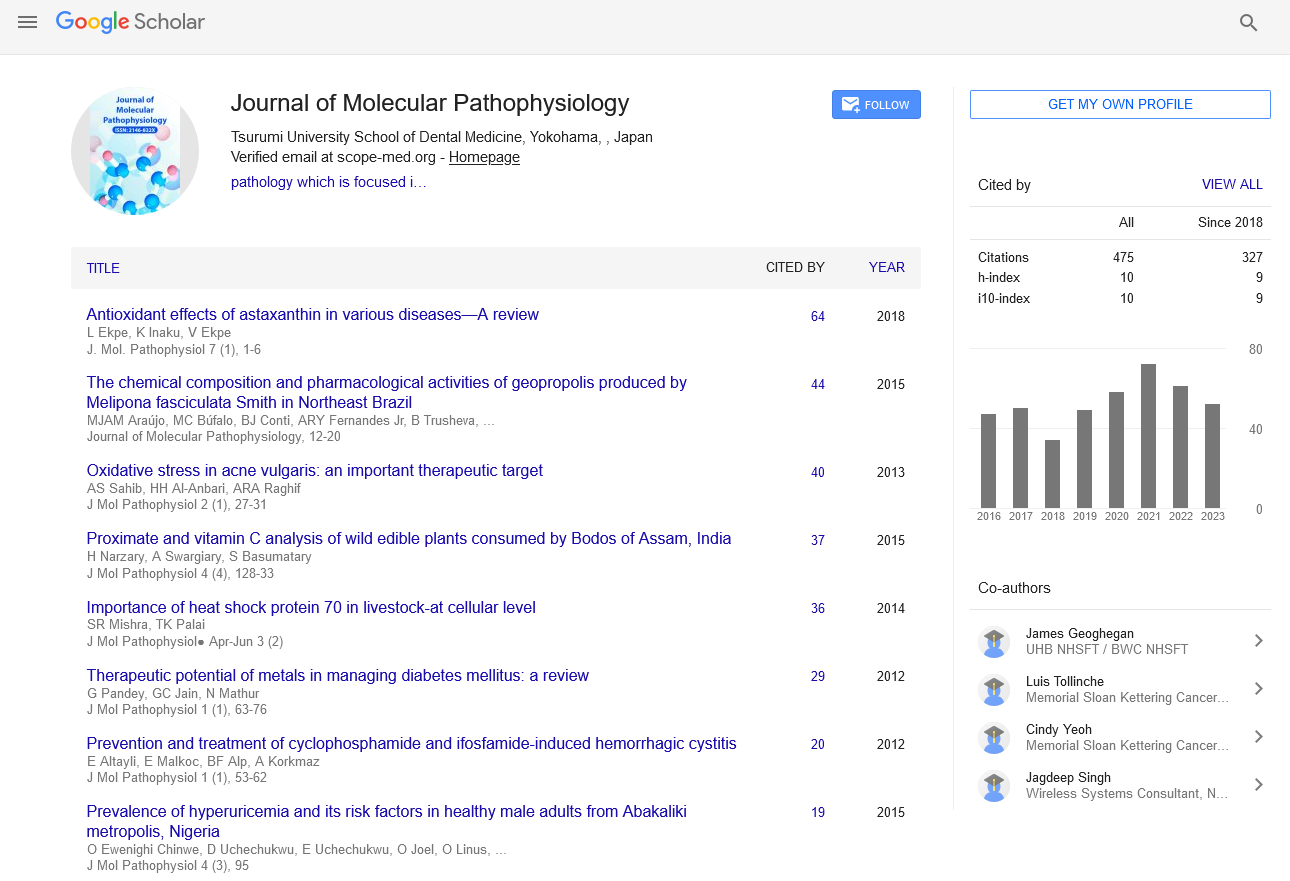
Opinion - Journal of Molecular Pathophysiology (2022)
Molecular Basis and Mechanisms of Epigenetics
Lydiea Cheng*Lydiea Cheng, Department of Neurosurgery, Medical University of Gdansk, Gdansk, Poland, Email: cheeng123@gmail.com
Received: 12-Sep-2022, Manuscript No. JMOLPAT-22-80571; Editor assigned: 15-Sep-2022, Pre QC No. JMOLPAT-22-80571 (PQ); Reviewed: 29-Sep-2022, QC No. JMOLPAT-22-80571; Revised: 07-Oct-2022, Manuscript No. JMOLPAT-22-80571 (R); Published: 14-Oct-2022
Description
While epigenetics is crucial to eukaryotes, particularly metazoans, bacteria use it differently from eukaryotes. Most notably, eukaryotes regulate gene expression predominantly through epigenetic processes, unlike bacteria. However, postreplicative DNA methylation is frequently employed by bacteria for the epigenetic regulation of DNA-protein interactions. Additionally, bacteria use DNA adenine methylation as an epigenetic signal rather than DNA cytosine methylation. Bacterial pathogenicity in species including Escherichia coli, Salmonella, Vibrio, Yersinia, Haemophilus, and Brucella depends on DNA adenine methylation. Adenine’s methylation in alphaproteobacteria controls the cell cycle and links DNA replication and gene transcription. Adenine methylation in gammaproteobacteria provides cues for DNA replication, chromosome segregation, mismatch repair, bacteriophage packaging, transposase activity, and gene expression control. Streptococcus pneumoniae (the pneumococcus) has a genetic switch that enables the bacteria to randomly change its features into six distinct states that may help develop better vaccinations. An automated phase variable methylation method generates each form at random. In each of these six phases, the pneumococcus has a varied potential for causing lethal infections. In other bacterial genera, there are comparable systems. Adenine methylation controls host adaptability, biofilm development, and sporulation in Bacillota like Clostridioides difficile.
Mechanisms
Epigenetic pathways play a significant role in determining the adult phenotype because they are responsive to environmental influences. Cell memory has come to be seen as involving a variety of epigenetic inheritance processes; however, not all of these are generally acknowledged as epigenetic in nature. An essential component of the evolutionary origin of cell differentiation was epigenetic processes. There have been some findings of transgenerational epigenetic inheritance, despite the fact that epigenetics in multicellular organisms is typically assumed to be a mechanism involved in differentiation, with epigenetic patterns “reset” when organisms reproduce. There is still a chance that multigenerational epigenetics could be an additional factor in evolution and adaptation, even if the majority of these features are gradually lost over multiple generations. Some people define epigenetics as heritable, as was mentioned above. Predetermined and probabilistic epigenesis is two categories of developmental epigenetics. Predetermined epigenesis is a one-way transition from DNA structure development to protein functional maturation. Predetermined here refers to scripted and predictable growth. On the other hand, probabilistic epigenesis is a bidirectional structure-function development that takes experiences and external moulding into account.
Molecular basis
Changes in the epigenome affect which genes are activated, but not the DNA’s genetic coding. Modifications to the chromatin proteins or DNA’s own architecture may activate or silence cells. A multicellular organism’s differentiated cells can express only the genes required for their own activities thanks to this technique. Cell division does not erase epigenetic modifications. The majority of epigenetic changes only take place during one organism’s lifespan; however, through a process known as transgenerational epigenetic inheritance, certain epigenetic alterations can be passed on to the organism’s children. Additionally, this epigenetic change might potentially be passed down to the following generation if gene inactivation takes place in a sperm or egg cell that leads to fertilisation. Paramutation, bookmarking, imprinting, gene silencing, X chromosome inactivation, position effect, DNA methylation reprogramming, transection, maternal effects, the development of carcinogenesis, numerous teratogen effects, regulation of histone modifications and heterochromatin, and technical restrictions affecting parthenogenesis and cloning are examples of specific epigenetic processes.
Copyright: © 2022 The Authors. This is an open access article under the terms of the Creative Commons Attribution NonCommercial ShareAlike 4.0 (https://creativecommons.org/licenses/by-nc-sa/4.0/). This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.